New Computational Approach Improves on Identification of Biomarkers
Recent work by a consortium of researchers demonstrates that a novel, network-based, analytical approach is effective in identifying predictive biomarkers of chemotherapy response in triple-negative breast cancer (TNBC) patient-derived xenografts (PDX) models. An update to this tool was presented at the 2024 AACR annual meeting.
The work was led by a team at Baylor College of Medicine, who recently published the paper, Identifying biomarkers of differential chemotherapy response in TNBC patient-derived xenografts with a CTD/WGCNA approach, in iScience. Their work demonstrates a way to more accurately accelerate testing of therapeutics to inform human clinical trials.
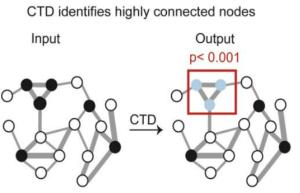
“Imagine one TNBC gene as a highly expressed ‘influencer’ relative to other genes that may be ‘quieter’ in terms of their expression,” said Aleksandar Milosavljevic, PhD, Henry and Emma Meyer Chair in Molecular Genetics and Director, Quantitative & Computational Biosciences, Baylor College of Medicine. “We show that we can more precisely identify specific genes as predictors of therapy response that otherwise might be missed through other clustering computational approaches, notably weighted gene co-expression network analysis (WCGNA). This work shows that the network-based approach called Connect the Dots (CTD) in combination with WGCNA is better at identifying quieter but relevant genes than WGCNA alone.”
In setting the stage for demonstrating the predictive value of genetic mutations in the TNBC PDX models they used, the team demonstrated that these models have a spectrum of gene mutations similar to the spectrum of human mutations in TNBC, based on comparative analysis using NCI’s The Cancer Genome Atlas (TCGA) data, accessible through the CRDC. Additionally, based on proteomic data available through the NCI’s Clinical Proteomic Tumor Analysis Consortium (CPTAC), the PDX models were shown to have clustering patterns similar to human TNBC proteomic profiles. CPTAC data are also accessible through the CRDC.
Finally, the team showed that responses in the TNBC PDX models to human-equivalent doses of docetaxel are in concord with the response of the tumor-of-origin when challenged with taxane. Results from one PDX sample show that platinum-containing regimens may be concordant to tumor of origin response. Both of these drugs are frequently used in human TNBC treatment, alone or in combination.
“This computational approach shows promise in identifying predictors of drug response in TNBC PDX models that may translate to human trials and eventually to more targeted patient care,” said Varduhi Petrosyan, PhD, lead author and graduate student in Molecular and Human Genetics, Baylor College of Medicine.
The new tool was developed with support from a team with Seven Bridges Cancer Genomics Cloud powered by Velsera, one of three NCI Cloud Resources. A second generation of the tool, called CTD2, was presented at a 2024 AACR annual meeting poster session. CTD2 has been refactored to dramatically increase its speed on the platform, and an additional algorithm called ‘Guilt by Association’ has been added to identify markers in the gene network that may be related to disease process but that are not surfaced by other analyses. CTD2 will be released to the public later this year.
A webinar on the CTD tool (first generation) can be found here.